Wrapper function for desplot::desplot() across multiple columns
desplot_across.RdThe goal of this function is to allow the user a quick and easy glance at the experimental layout of a trial by creating multiple desplots at once - one for each variable that is provided (see example below).
Arguments
- data
A data frame.
- vars
Vector with variables/column names for which a desplot should be created.
- formright
A formula like
x*y|location, i.e. the right-hand side of the formulayield~x*y|locationthat would usually be passed todesplot::desplot(form = ...). Note thatxandyare numeric and the default is"col + row".- lang
Language for plots labels.
- title
Can either be
"none","short"or"long". For the respectivevar, it gives information about the number of levels and their respective frequency in the data.- flip
see
desplot::desplot()documentation - set to opinionated default here- out1.gpar
see
desplot::desplot()documentation - set to opinionated default here- out2.gpar
see
desplot::desplot()documentation - set to opinionated default here- ...
Other arguments passed to
desplot::desplot().
Value
A list of desplots.
Examples
library(BioMathR)
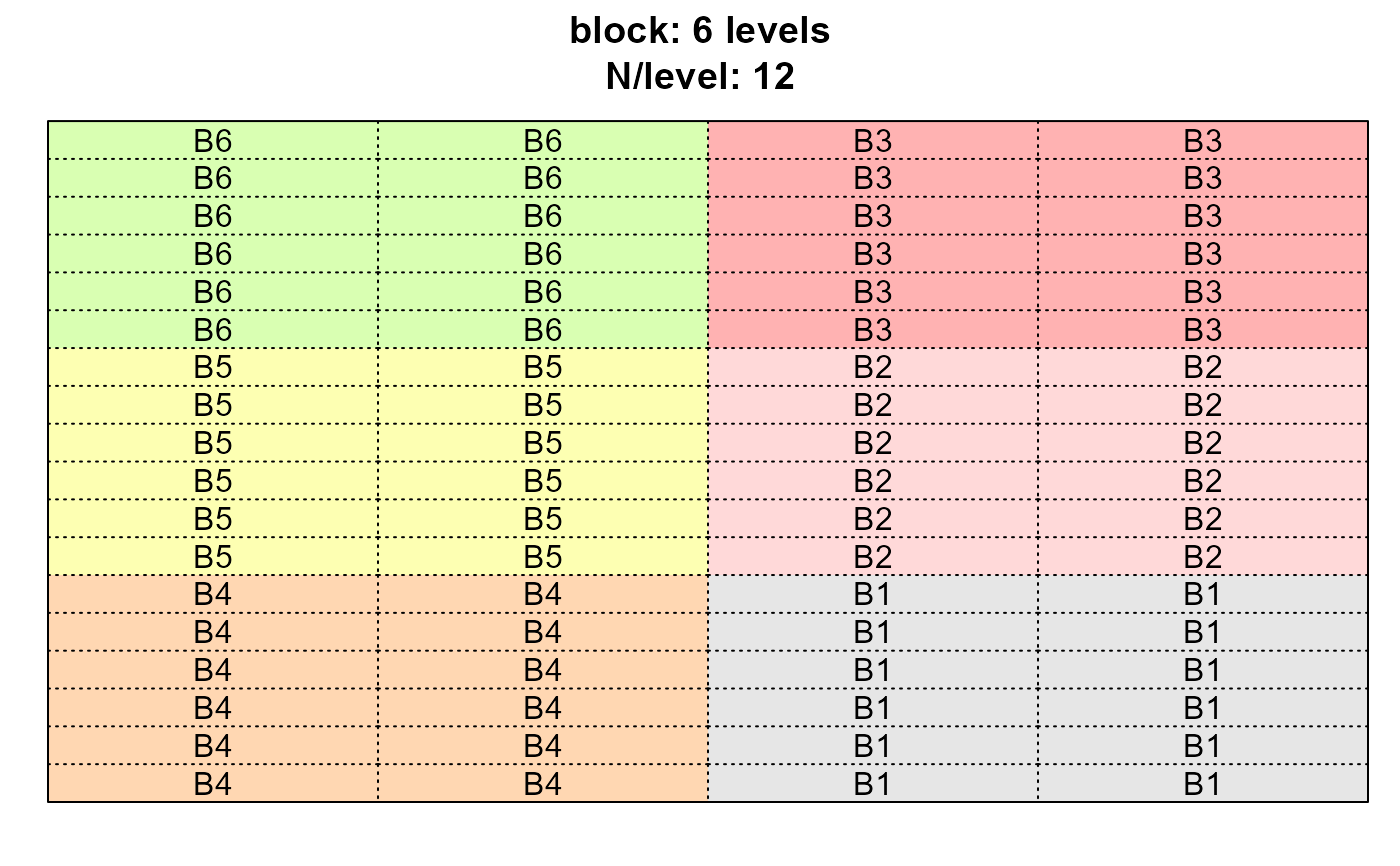
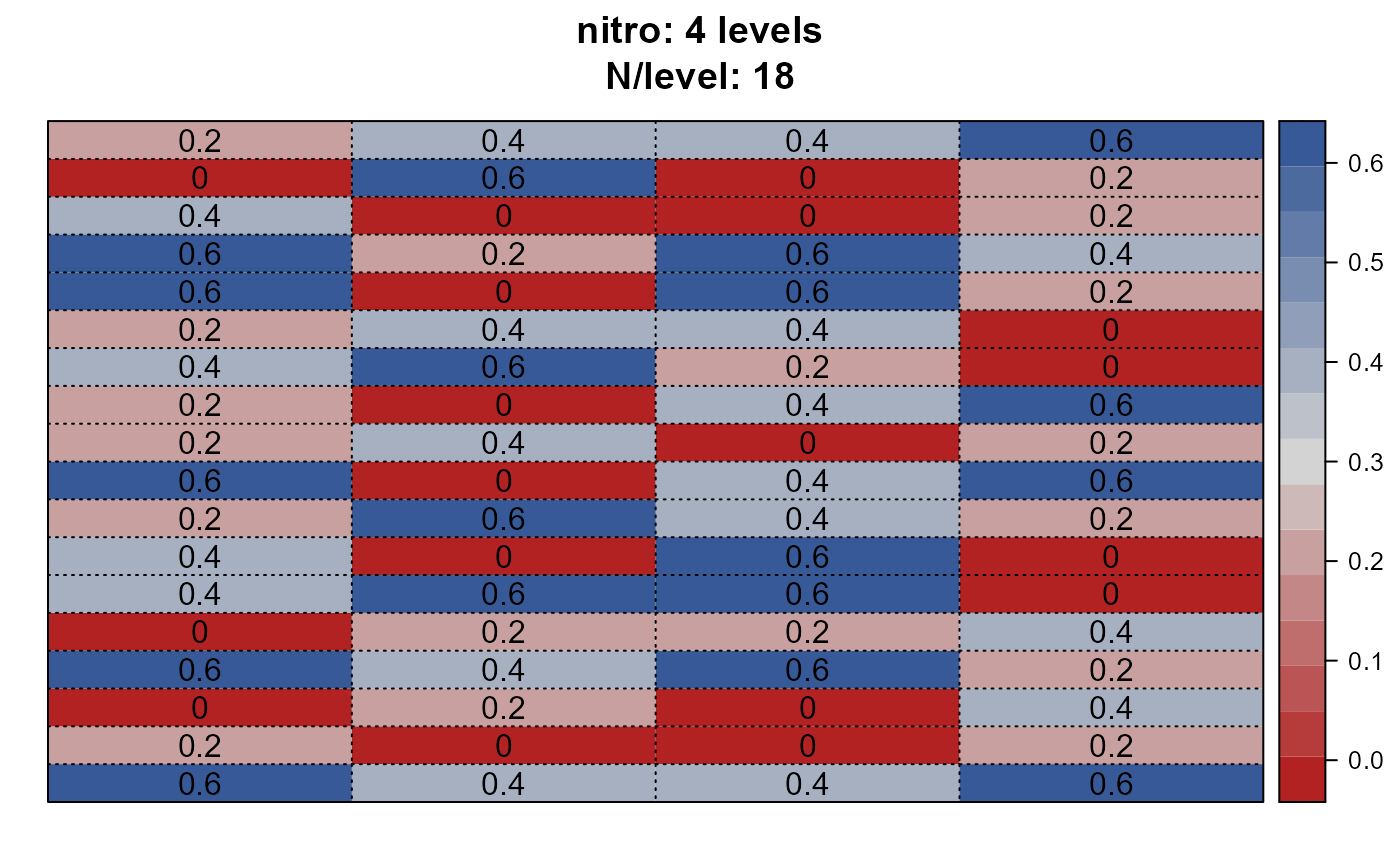
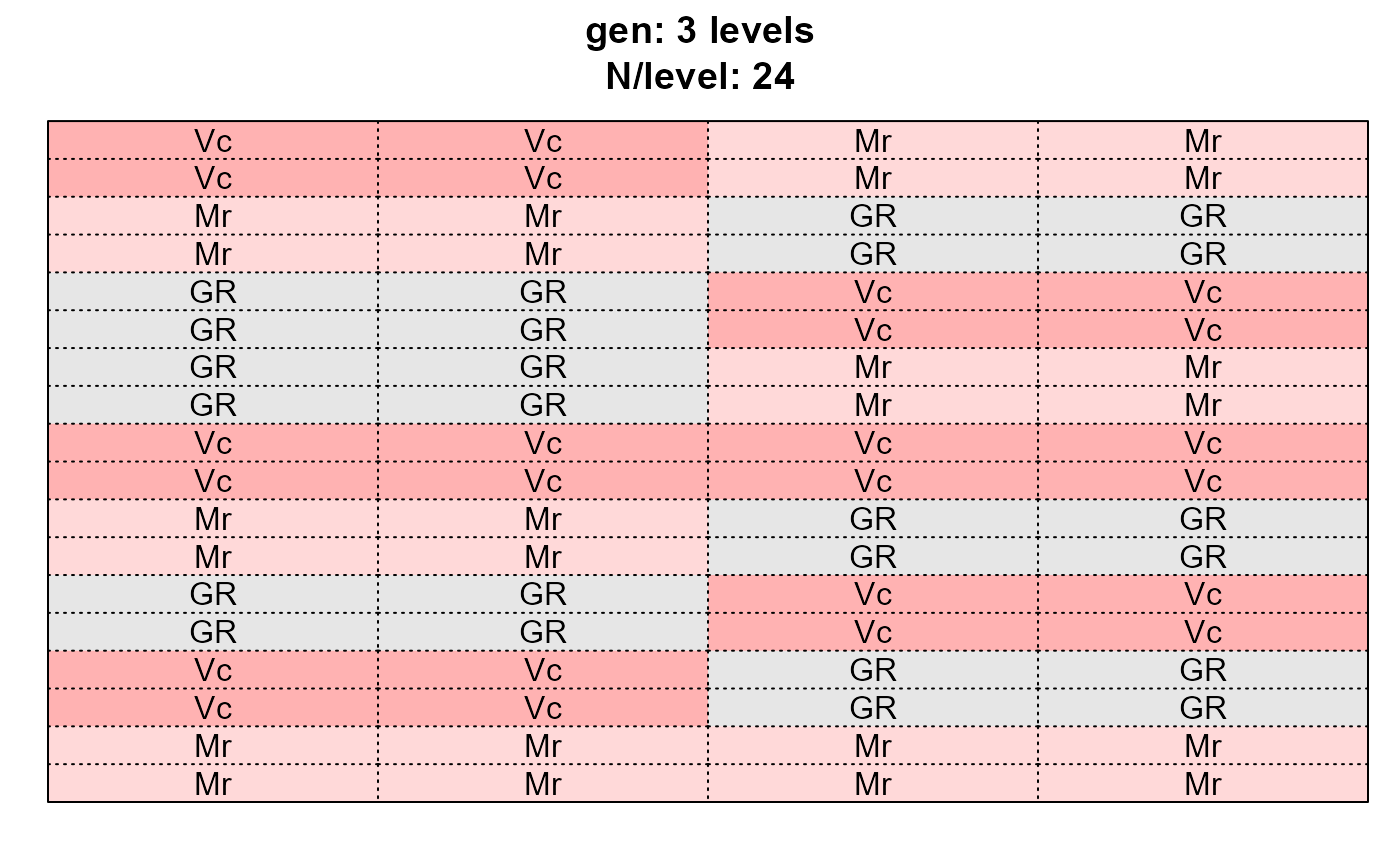
dps <- desplot_across(data = agridat::yates.oats,
vars = c("nitro", "gen", "block"),
cex = 1)
dps$nitro
 dps$gen
dps$gen
 dps$block
dps$block